 PDF(2833 KB)
PDF(2833 KB)


云南省德宏州东方库蠓中西藏环状病毒的分离鉴定
杨怡铃, 何于雯, 李楠, 孟锦昕, 殷建忠, 王静林
中国媒介生物学及控制杂志 ›› 2022, Vol. 33 ›› Issue (4) : 580-585.
 PDF(2833 KB)
PDF(2833 KB)
 PDF(2833 KB)
PDF(2833 KB)
云南省德宏州东方库蠓中西藏环状病毒的分离鉴定
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Isolation and identification of Tibet orbivirus from Culicoides orientalis collected in Dehong prefecture, Yunnan province, China
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}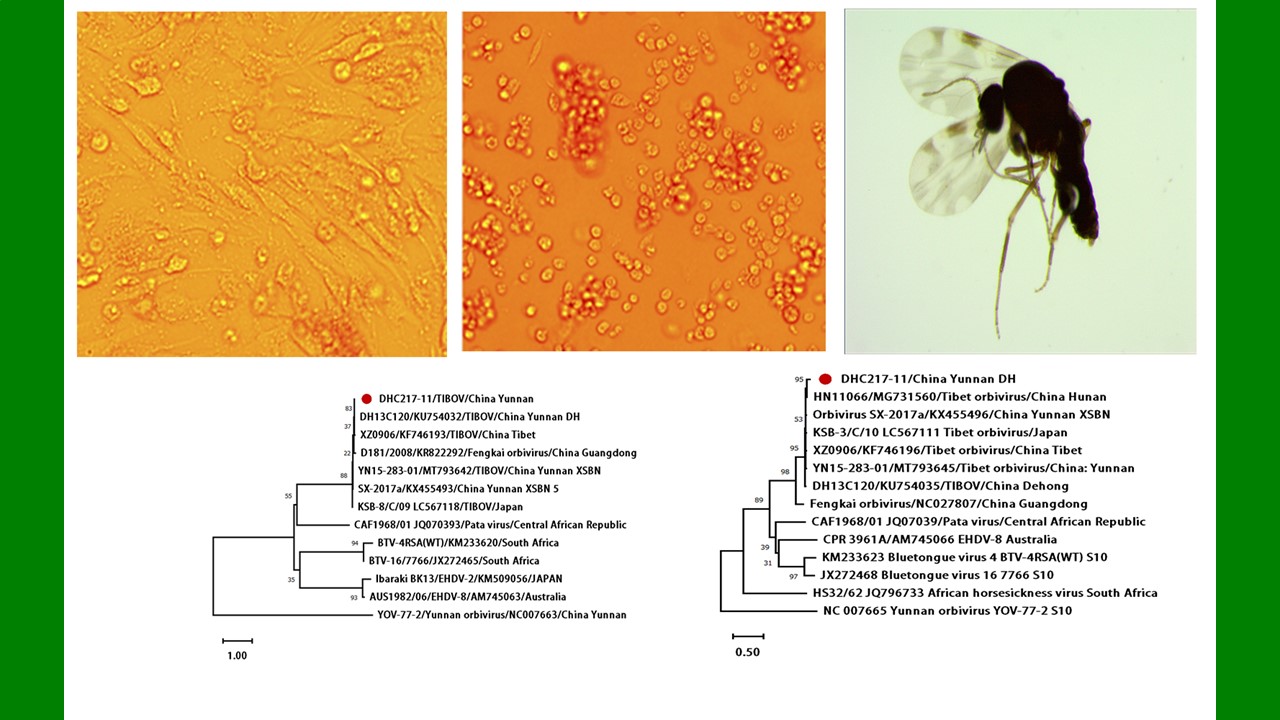
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |