 PDF(3315 KB)
PDF(3315 KB)


范张食酪螨和腐食酪螨的形态学与分子生物学鉴定
范笑忱, 刘璐瑶, 方瑜, 褚凌渺, 冯蕊, 李飞燕, 潘若兮, 王少圣, 孙恩涛
中国媒介生物学及控制杂志 ›› 2022, Vol. 33 ›› Issue (2) : 277-280.
 PDF(3315 KB)
PDF(3315 KB)
 PDF(3315 KB)
PDF(3315 KB)
范张食酪螨和腐食酪螨的形态学与分子生物学鉴定
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Morphological and molecular biological identification of Tyrophagus fanetzhangorum and T. putrescentiae
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}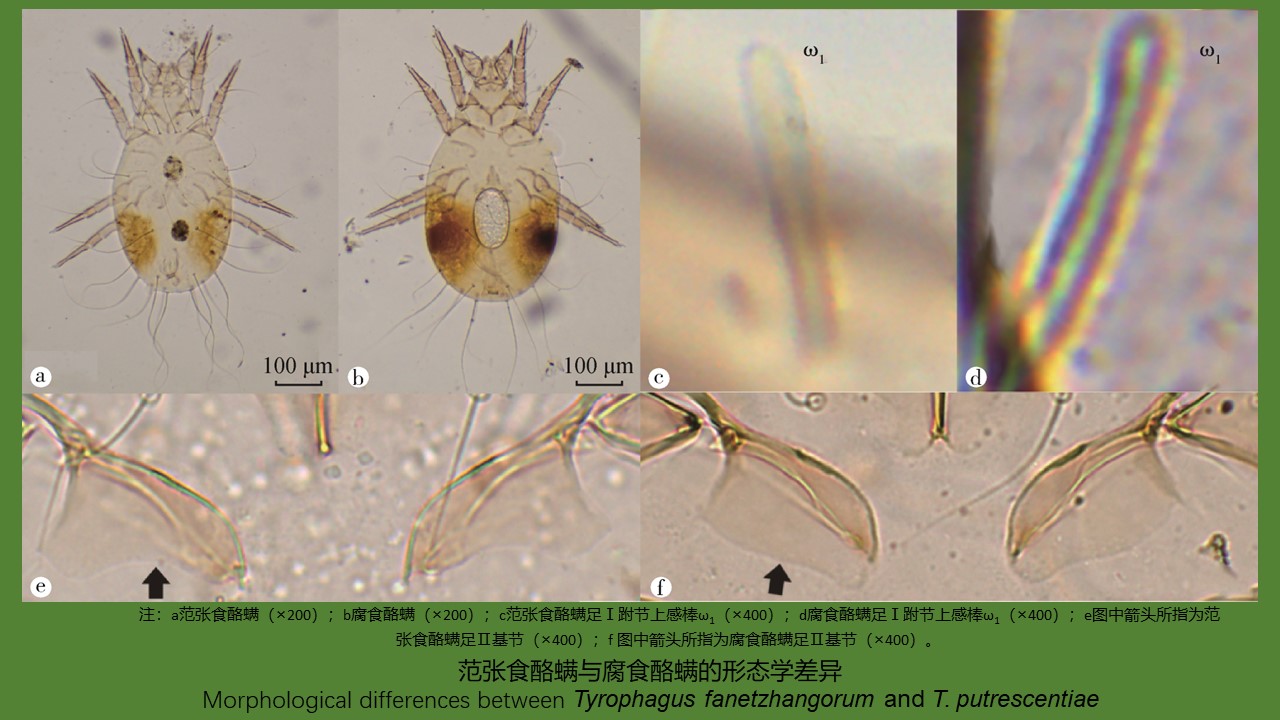
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |