 PDF(818 KB)
PDF(818 KB)


基于4种线粒体基因序列的广东省南雄市农区小型兽类DNA条形码分析
姜洪雪, 姚丹丹, 林思亮, 冯志勇
中国媒介生物学及控制杂志 ›› 2022, Vol. 33 ›› Issue (1) : 48-53.
 PDF(818 KB)
PDF(818 KB)
 PDF(818 KB)
PDF(818 KB)
基于4种线粒体基因序列的广东省南雄市农区小型兽类DNA条形码分析
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}DNA barcoding analysis of small mammals in the agricultural area in Nanxiong of Guangdong province based on four mitochondrial gene sequences
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}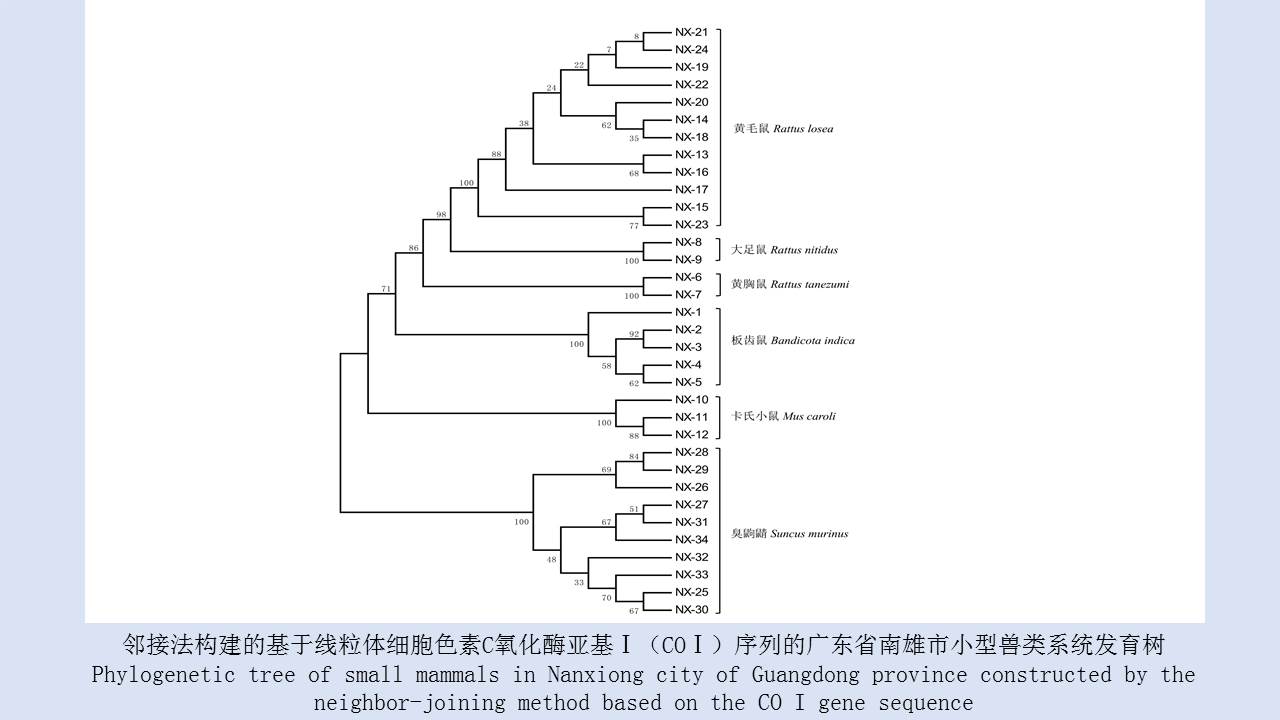
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |