 PDF(1483 KB)
PDF(1483 KB)


我国部分地区蜱体弗朗西斯菌属和弗类共生体的检测研究
王艳华, 孙英伟, 毛玲玲, 王子江, 张家勇, 张稷博, 彭遥, 夏连续
中国媒介生物学及控制杂志 ›› 2021, Vol. 32 ›› Issue (6) : 756-762.
 PDF(1483 KB)
PDF(1483 KB)
 PDF(1483 KB)
PDF(1483 KB)
我国部分地区蜱体弗朗西斯菌属和弗类共生体的检测研究
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Detection of Francisella strains and Francisella-like endosymbionts in ticks in some regions of China
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}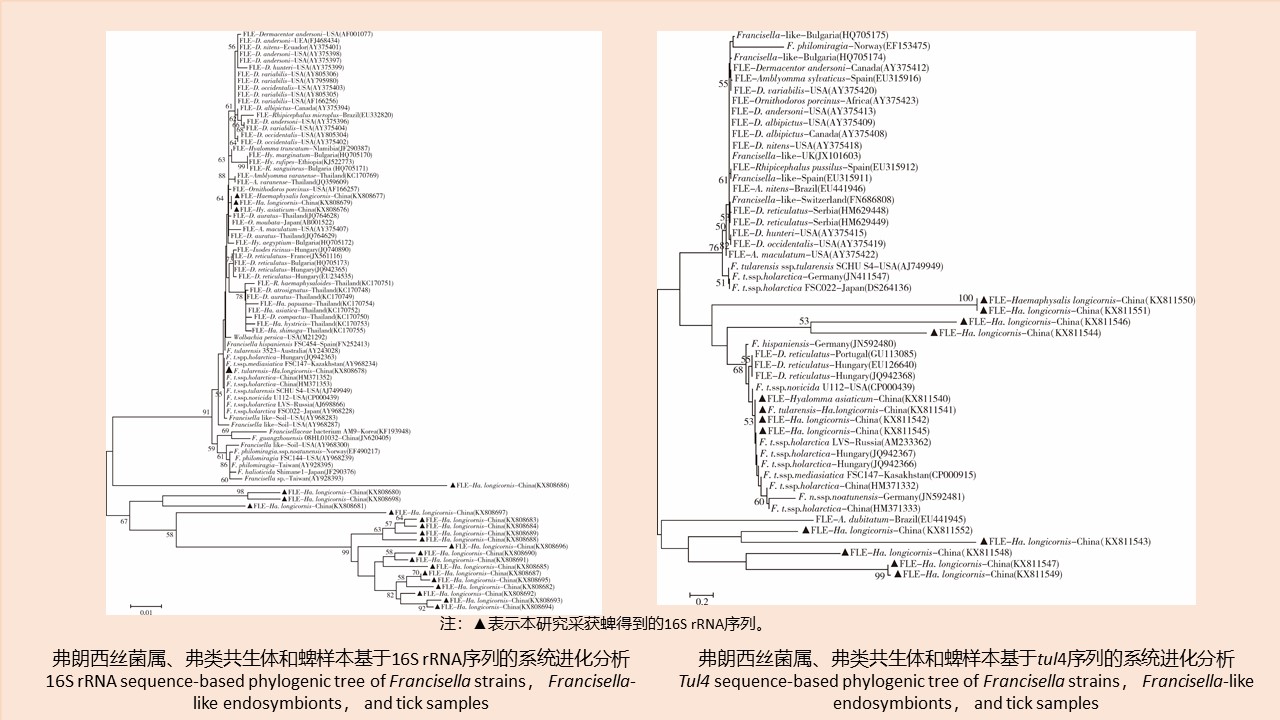
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |