 PDF(1054 KB)
PDF(1054 KB)


基于多态微卫星DNA的中国雷氏按蚊群体遗传结构研究
周秋明, 林琳, 董昊炜, 袁浩, 白洁, 李翔宇, 彭恒, 马雅军
中国媒介生物学及控制杂志 ›› 2021, Vol. 32 ›› Issue (5) : 526-532.
 PDF(1054 KB)
PDF(1054 KB)
 PDF(1054 KB)
PDF(1054 KB)
基于多态微卫星DNA的中国雷氏按蚊群体遗传结构研究
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Genetic structure of Anopheles lesteri populations in China based on microsatellite loci
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}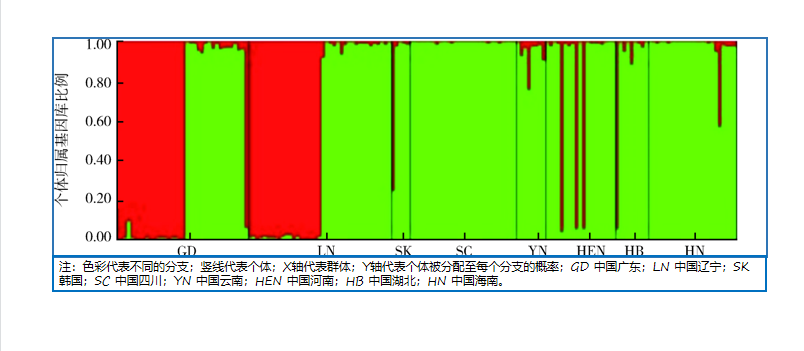
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |