 PDF(2076 KB)
PDF(2076 KB)


基于mtDNA-COⅠ基因的山东省淡色库蚊种群遗传多样性分析
臧传慧, 李黎明, 刘烁, 公茂庆, 王文倩, 王奕婷, 娄紫薇, 类晶晶, 程鹏, 刘宏美
中国媒介生物学及控制杂志 ›› 2023, Vol. 34 ›› Issue (3) : 298-302.
 PDF(2076 KB)
PDF(2076 KB)
 PDF(2076 KB)
PDF(2076 KB)
基于mtDNA-COⅠ基因的山东省淡色库蚊种群遗传多样性分析
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Genetic diversity of Culex pipiens pallens populations in Shandong province, China based on the mtDNA-COⅠgene
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}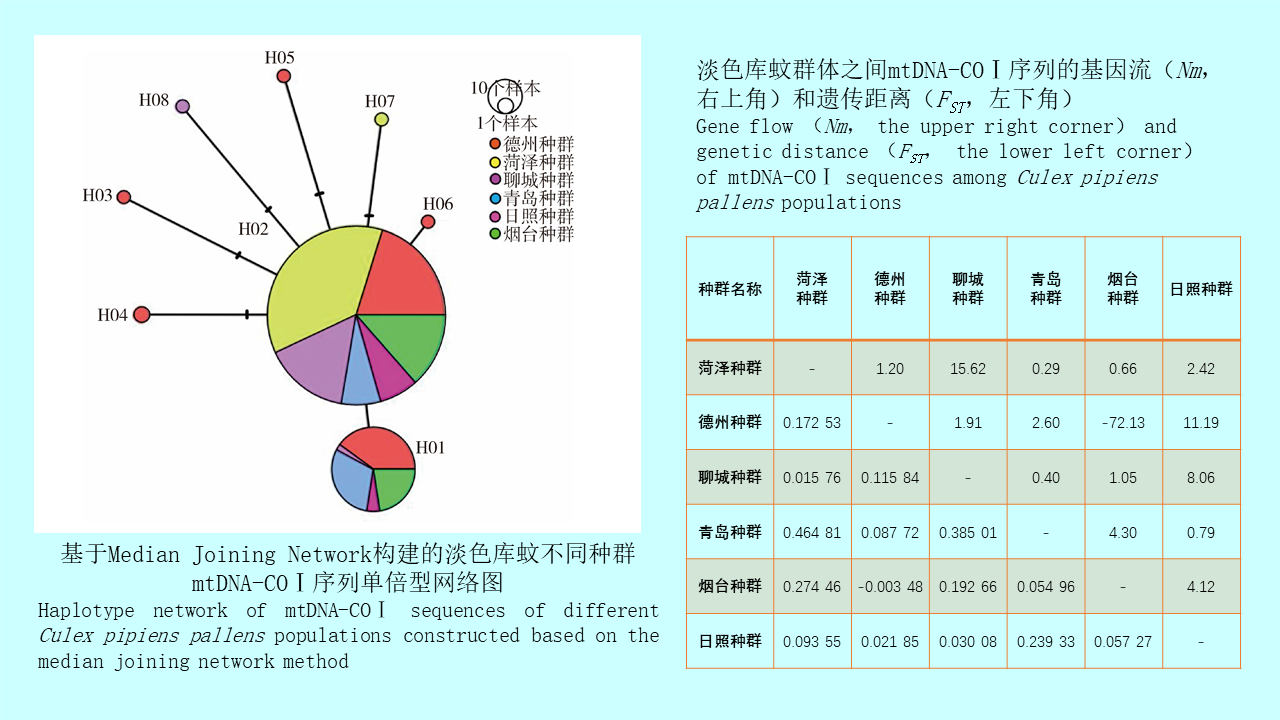
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |