 PDF(1326 KB)
PDF(1326 KB)


内蒙古自治区3个地区布氏田鼠的种群遗传多样性及种群遗传结构研究
刘雨秋, 鲁亮, 刘蓬勃, 赵宁, 李贵昌, 栗冬梅, 宋秀平, 王君, 刘起勇
中国媒介生物学及控制杂志 ›› 2023, Vol. 34 ›› Issue (3) : 291-297.
 PDF(1326 KB)
PDF(1326 KB)
 PDF(1326 KB)
PDF(1326 KB)
内蒙古自治区3个地区布氏田鼠的种群遗传多样性及种群遗传结构研究
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}Genetic diversity and genetic structure of Lasiopodomys brandtii populations in three regions of Inner Mongolia, China
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}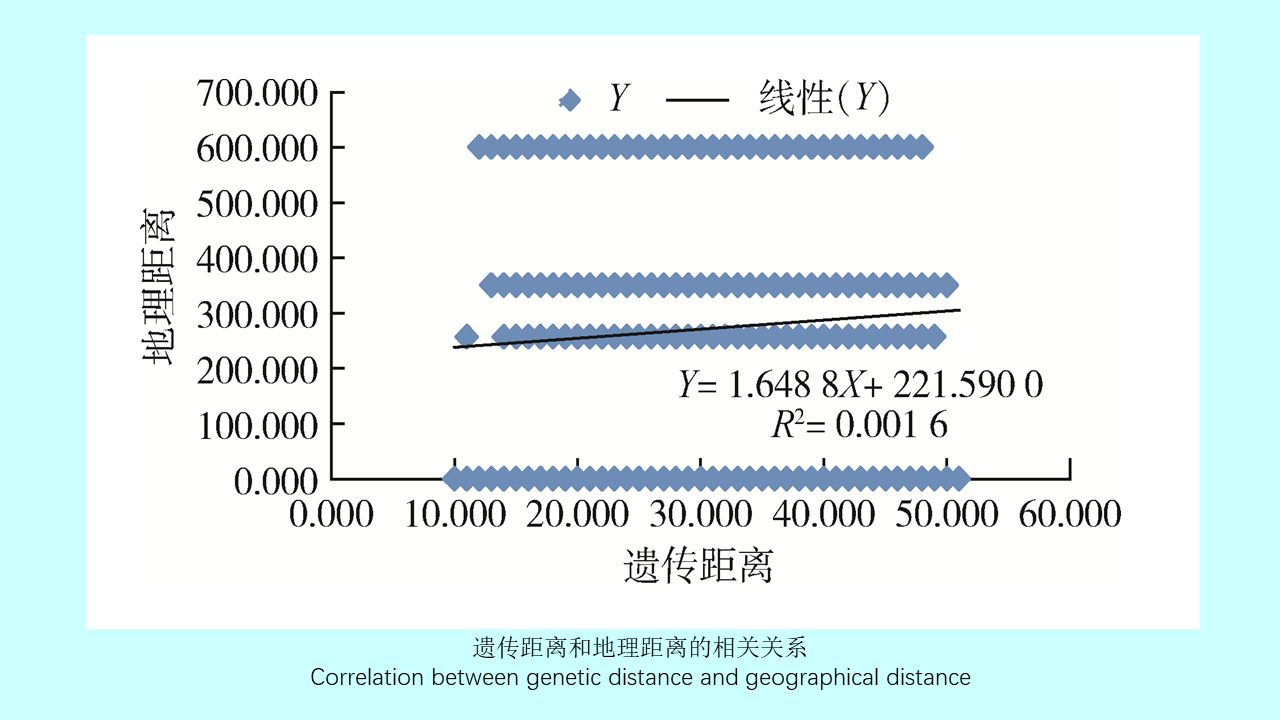
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |