 PDF(1742 KB)
PDF(1742 KB)


白纹伊蚊昌平种群高效氯氰菊酯抗性的比较蛋白质组学研究
华栋栋, 王磊, 肖迪, 李文玉, 周欣欣, 伦辛畅, 母群征, 刘起勇, 马伟, 孟凤霞
中国媒介生物学及控制杂志 ›› 2023, Vol. 34 ›› Issue (2) : 196-203.
 PDF(1742 KB)
PDF(1742 KB)
 PDF(1742 KB)
PDF(1742 KB)
白纹伊蚊昌平种群高效氯氰菊酯抗性的比较蛋白质组学研究
 ({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}
({{custom_author.role_cn}}), {{javascript:window.custom_author_cn_index++;}}A comparative proteomics analysis of beta-cypermethrin resistance in Aedes albopictus population in Changping, Beijing, China
 ({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}
({{custom_author.role_en}}), {{javascript:window.custom_author_en_index++;}}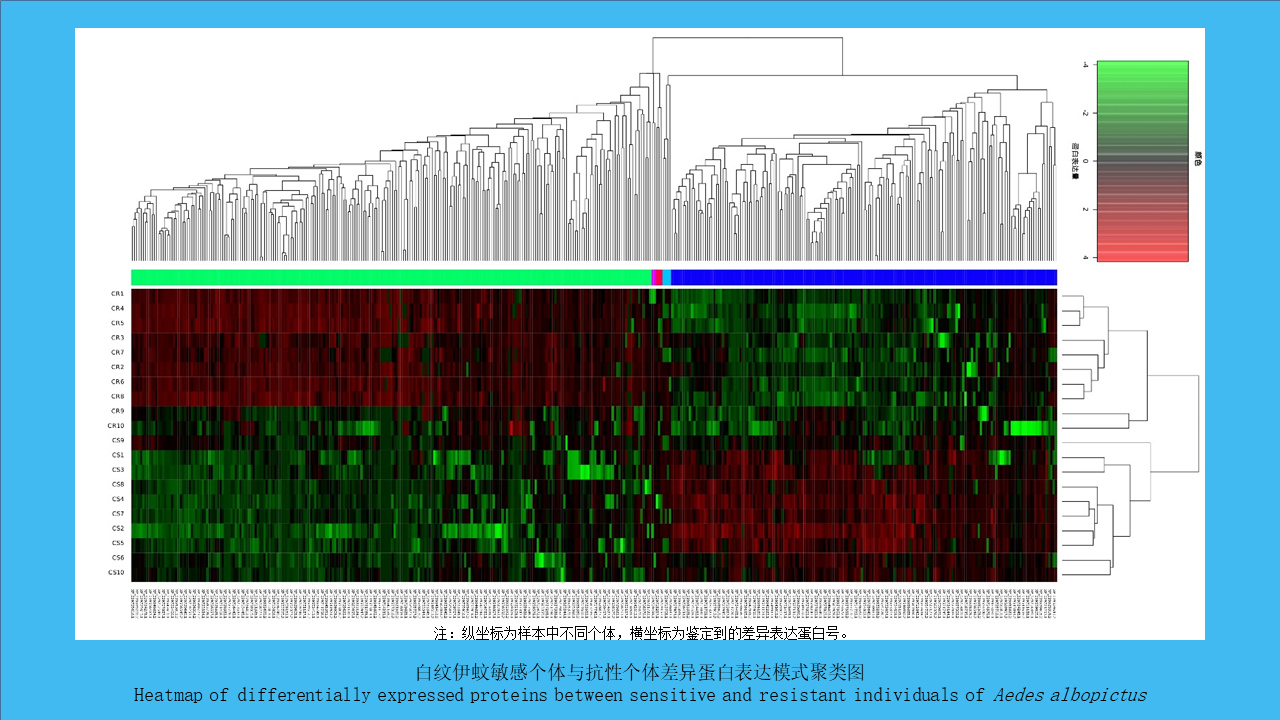
| {{custom_ref.label}} |
{{custom_citation.content}}
{{custom_citation.annotation}}
|
/
| 〈 |
|
〉 |