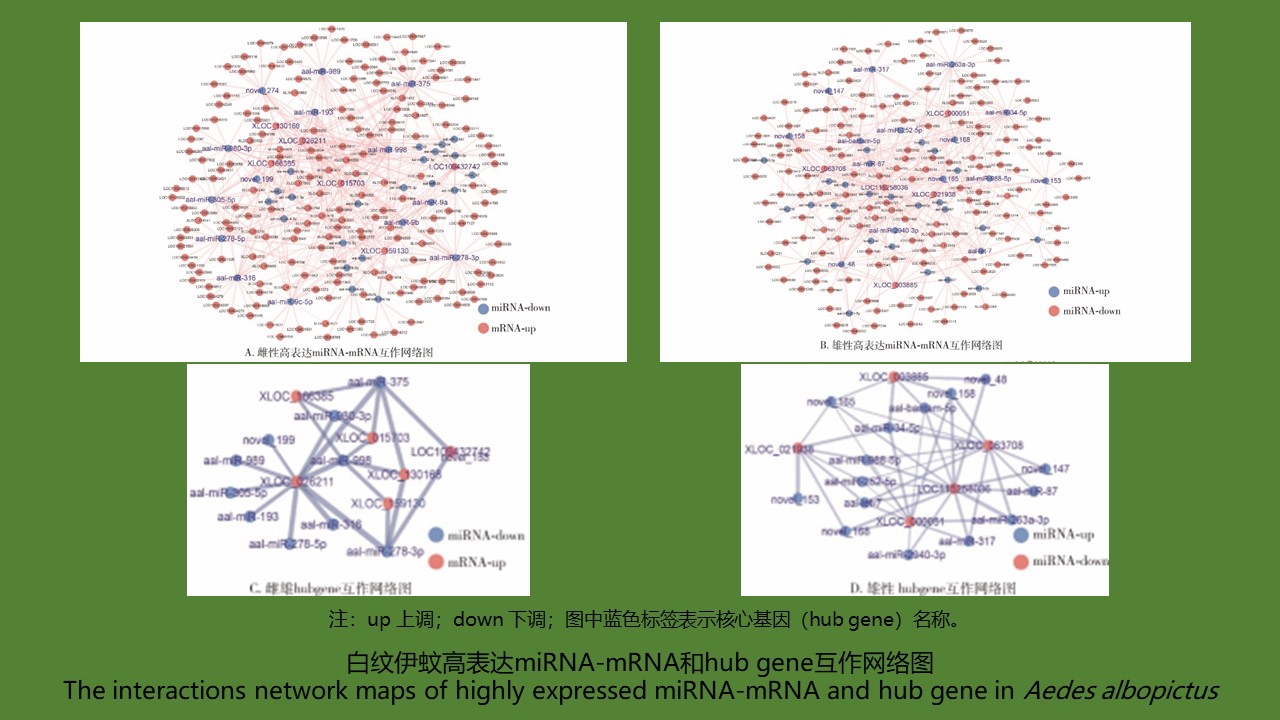
Objective To investigate differentially expressed miRNAs between male and female Aedes albopictus and their target mRNAs,explore miRNA-mRNA regulatory relationship in male and female Ae. albopictus,and predict the potential function of key genes, and to provide evidence for clarifying the regulatory mechanisms of sex differentiation of Ae. albopictus. Methods Through small RNA library construction and mRNA sequencing analysis,differentially expressed miRNAs and mRNAs between female and male Ae. albopictus were identified. Bioinformatic tools were used to predict the target mRNAs of the differentially expressed miRNAs. The miRNA-mRNA interaction networks were constructed using the overlapping results between the predicted target mRNAs and the sex differentially expressed mRNAs. GO and KEGG enrichment analysis were performed on the target mRNAs of miRNAs to predict the possible functions of miRNAs. Results A total of 74 sex differentially expressed miRNAs were screened out,including 8 sex-specific miRNAs. A total of 470 female-specific miRNA-mRNA pairs (down-regulated miRNAs,up-regulated mRNAs) and 481 male-specific miRNA-mRNA pairs (up-regulated miRNAs,down-regulated mRNAs) were acquired. The interaction network and functional enrichment analyses determined 4 mRNAs associated with the sex differentiation and reproduction of Ae. albopictus and 17 miRNAs that directly regulated these mRNAs. Conclusion We constructed sex differentially expressed miRNA-mRNA interaction networks of Ae. albopictus,and acquired 17 miRNAs and 4 mRNAs that were related to sex differentiation, laying the foundation for further research on the role of miRNAs in regulating the sex differentiation of Ae. albopictus.
LIU Wen-juan, ZHANG Ke-xin, CHENG Peng, ZHANG Xin-yu, ZHANG Qian, ZHANG Zhong, ZHANG Rui-ling
. Deciphering sex differentially expressed miRNAs and miRNA-mRNA regulatory networks of Aedes albopictus[J]. Chinese Journal of Vector Biology and Control, 2022
, 33(2)
: 191
-200
.
DOI: 10.11853/j.issn.1003.8280.2022.02.005
[1] Franklinos LHV, Jones KE, Redding DW, et al. The effect of global change on mosquito-borne disease[J]. Lancet Infect Dis, 2019, 19(9):e302-e312. DOI:10.1016/S1473-3099(19)30161-6.
[2] 杨舒然, 刘起勇. 白纹伊蚊的全球分布及扩散趋势[J]. 中国媒介生物学及控制杂志, 2013, 24(1):1-4. Yang SR, Liu QY. Trend in global distribution and spread of Aedes albopictus[J]. Chin J Vector Biol Control, 2013, 24(1):1-4.(in Chinese)
[3] Palatini U, Masri RA, Cosme LV, et al. Improved reference genome of the arboviral vector Aedes albopictus[J]. Genome Biol, 2020, 21(1):215. DOI:10.1186/s13059-020-02141-w.
[4] 张强. 桔小实蝇发育阶段转变前后miRNA分析及miR-309a调控卵巢发育的分子机制[D]. 重庆:西南大学, 2020. DOI:10.27684/d.cnki.gxndx.2020.000080. Zhang Q. miRNA analysis of transitions between different developmental stages and the molecular mechanism of miR-309a regulating the ovarian development in Bactrocera dorsalis[D]. Chongqing:Southwest University, 2020. DOI:10.27684/d.cnki.gxndx.2020.000080.(in Chinese)
[5] Bushati N, Cohen SM. microRNA functions[J]. Annu Rev Cell Dev Biol, 2007, 23:175-205. DOI:10.1146/annurev.cellbio.23. 090506.123406.
[6] 赵星, 左丽. 伊蚊对登革病毒的垂直与水平传播[J]. 国外医学•流行病学传染病学分册, 2004, 31(2):108-110. DOI:10.3760/cma.j.issn.1673-4149.2004.02.015. Zhao X, Zuo L. Vertical and horizontal transmission of dengue virus by Aedes[J]. Epidemiol Lemol Foreign Med Sci, 2004, 31(2):108-110. DOI:10.3760/cma.j.issn.1673-4149.2004.02.015.(in Chinese)
[7] Parkhomchuk D, Borodina T, Amstislavskiy V, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA[J]. Nucleic Acids Res, 2009, 37(18):e123. DOI:10.1093/nar/gkp596.
[8] Zhou L, Chen JH, Li ZZ, et al. Integrated profiling of microRNAs and mRNAs:MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma[J]. PLoS One, 2010, 5(12):e15224. DOI:10.1371/journal.pone.0015224.
[9] Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12):550. DOI:10.1186/s13059-014-0550-8.
[10] Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila[J]. Genome Biol, 2003, 5(1):R1. DOI:10.1186/gb-2003-5-1-r1.
[11] Krüger J, Rehmsmeier M. RNAhybrid:MicroRNA target prediction easy, fast and flexible[J]. Nucleic Acids Res, 2006, 34(Suppl 2):W451-W454. DOI:10.1093/nar/gkl243.
[12] Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq:Accounting for selection bias[J]. Genome Biol, 2010, 11(2):R14. DOI:10.1186/gb-2010-11-2-r14.
[13] Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment[J]. Nucleic Acids Res, 2008, 36 Suppl 1:D480-D484. DOI:10.1093/nar/gkm882.
[14] Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J]. Genome Biol, 2009, 10(3):1-10. DOI:10.1186/gb-2009-10-3-r25.
[15] Wen M, Shen Y, Shi SH, et al. miREvo:An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments[J]. BMC Bioinformatics, 2012, 13:140. DOI:10.1186/1471-2105-13-140.
[16] Friedländer MR, Mackowiak SD, Li N, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades[J]. Nucleic Acids Res, 2012, 40(1):37-52. DOI:10.1093/nar/gkr688.
[17] Poelchau MF, Reynolds JA, Denlinger DL, et al. A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation[J]. BMC Genomics, 2011, 12:619. DOI:10.1186/1471-2164-12-619.


